NKE knowledge page
About
Landing page for the sodium potassium pump NKE. Other name for these proteins: NKA, Na-K-ATPase.
NKE helps maintain resting membrane potential through helping establish the sodium and potassium concentration gradients across the membrane.
NKE is thought to be a heterodimer - made of the active alpha subunit protein, which catalyses the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This is assembled with the regulatory beta subunit.
Pictures
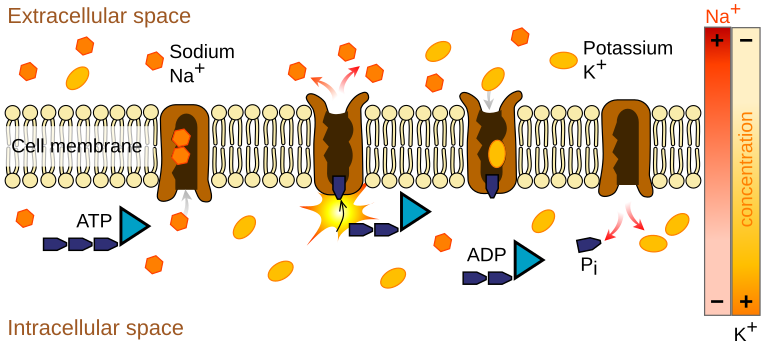
Subfamilies
The following list contains a portion of all subfamilies that can compose the NKE.
| Channel name | Alias | Gene | Structure |
|---|---|---|---|
| ATP1A1 | Uniprot A1 | PDB A1 | |
| ATP1A2 | Uniprot A2 | SMR A2 | |
| ATP1A3 | Uniprot A3 | PDB A3 | |
| ATP1A4 | Uniprot A4 | SMR A4 | |
| ATP1B1 | Uniprot B1 | PDB B1 | |
| ATP1B2 | Uniprot B2 | SMR B2 | |
| ATP1B3 | Uniprot B3 | SMR B3 | |
| ATP1C | ATNG, FXYD2 | Uniprot 1C | PDB 1C |
| FXYD3 | Uniprot FXYD3 | SMR FXYD3 |
Literature
| Title | Author |
|---|---|
| A computational model of induced pluripotent stem-cell derived cardiomyocytes incorporating experimental variability from multiple data sources | Kernik et al. |
| A thermodynamic framework for modelling membrane transporters | Pan et al. |
Existing models
| Title | Author |
|---|---|
| BG_NaK | S Fong |
| Cellular consequences of HERG mutations in the long QT syndrome: precursors to sudden cardiac death*^ | C Clancy and Y Rudy |
| Bond graph modelling of the cardiac action potential: Implications for drift and non-unique steady states^ | M Pan et al. |
| A Model of the Ventricular Cardiac Action Potential - Depolarisation, Repolarisation and Their Interaction*^ | C Luo and Y Rudy |
Models denoted by [*] are not in bond graph form. Models denoted by [^] are part of a whole cell model.
Other annotations
| Item | Database |
|---|---|
| Sodium:potassium-exchanging ATPase complex | Gene Ontology Complex |
| P-type sodium:potassium-exchanging transporter activity | Gene Ontology activity |

