This workspace describes my (David Nickerson) attempt to create a validated CellML implementation of the Cortassa et al (2006) excitation-contraction coupled with mitochondrial energetics ECME model from Biophys J (http://www.biophysj.org/cgi/content/abstract/91/4/1564). So far I have just extracted the subset of files needed to run the basic periodic stimulus simulation described in Figure S1 of the supplement to the Biophys J article. Furthermore, this version of the model is largely just an encoding of the model based on Sonia's original C-code with minimal effort to follow CellML 1.1 best practices, so there isn't much smarts in the CellML encoding. For example, a lot of the units are marked "TODO" and set to dimensionless.
The normative version of this model is the CellML 1.1 version found in the cellml folder of this workspace. Currently the primary simulation experiment simply applies the periodic stimulus as described in the original Figure S1. This experiment can be found here: cellml/experiments/S1.xml. As an aid to use of this model I have also added a CellML 1.0 version of the S1 experiment as cellml-1.0/S1.xml. The CellML 1.0 version was generated using Jonathan Cooper's VersionConverter utility. Both versions of these models seem to run in OpenCell (version 0.8) and produce qualitatively the same results (I haven't done a quantitative comparison).
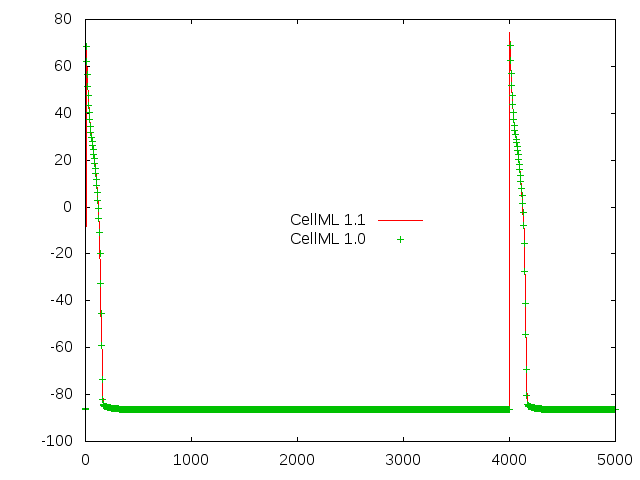
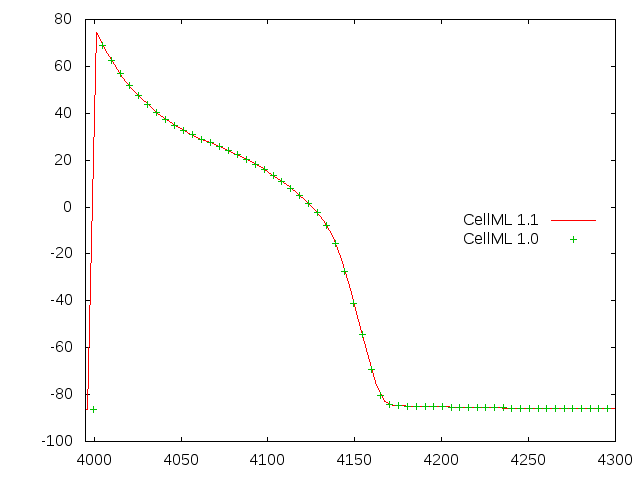
You can see that this encoding of the model gives reasonable looking action potentials as shown below. The two traces are for the normative CellML 1.1 version of the model and the generated CellML 1.0 version.

If you find any of these models useful in your work, please cite this exposure. If you would like to help develop this validated implementation of the model please contact me.