BG_cAMP
About this model
This is a bond-graph model of cyclic AMP (cAMP) and adenylyl cyclase (AC) metabolism in the cardiac cell.
- INPUTS:
- ATP stimulus
- OUTPUTS:
- Change in molar amounts of AC, cAMP
- REACTIONS:
- Re1: Conversion of ATP into cAMP via AC
- Re2: Conversion of ATP into cAMP via the activated alpha unit of the Gs protein bound by AC (GsαGTP:AC)
- Re3: Conversion of ATP into cAMP via the forskolin-AC complex (FSK:AC)
- Re4: Cleavage of cAMP into 5'AMP via a phosphodiesterase (PDE)
- Re5: Inhibition of PDE by IBMX
- Re6: Binding of GsαGTP to AC
- Re7: Binding of FSK to AC
- Re8: Inhibition of AC by the activated alpha unit of the Gi protein (GiαGTP)
Model status
The current CellML implementation runs in OpenCOR.
Model overview
This model is made from an existing kinetic model, where the mathematics are translated into the bond-graph formalism. This describes the model in energetic terms and forces adherence to the laws of thermodynamics.
Most reactions follow Michaelis-Menten kinetics, where an intermediate complex is made before the final product is created (e.g. Re:1a and Re:1b). All other reactions (Re5-8) follow classical mass-action kinetics.
For the following figure, all enzymes of a given reaction are shown in maroon.
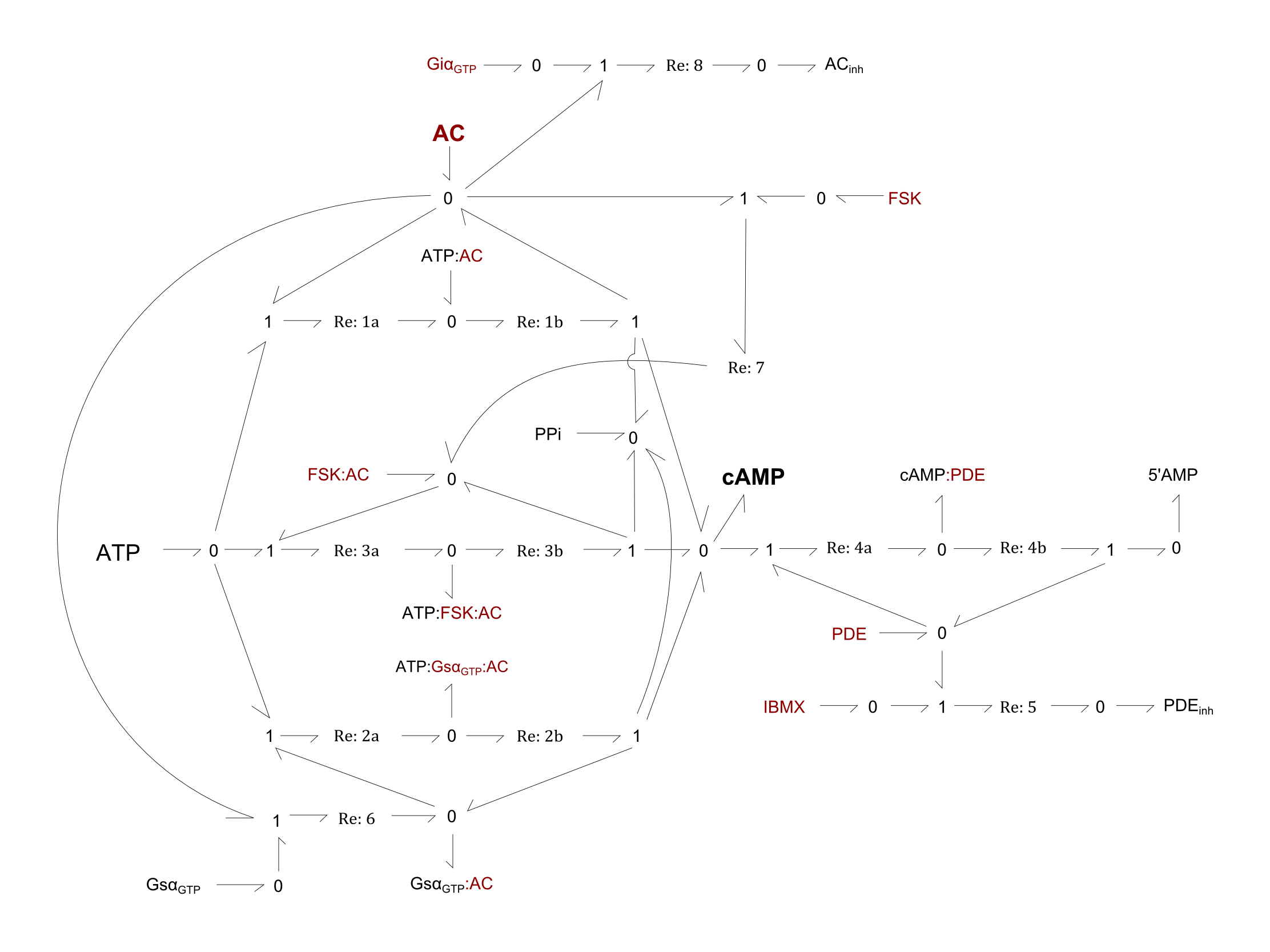
Fig. 1. Bond-graph formulation of the cAMP network
For the above bond-graphs, a '0' node refers to a junction where all chemical potentials are the same. A '1' node refers to all fluxes being the same going in and out of the junction.
| Abbreviation | Name |
|---|---|
| AC | Adenylyl cyclase |
| ACinh | Inactivated variant of adenylyl cyclase |
| ATP | Adenosine triphosphate |
| cAMP | cyclic AMP |
| FSK | forskolin |
| Giα | alpha subunit of the Gi protein |
| Gsα | alpha subunit of the Gs protein |
| IBMX | 3-isobutyl-1-methylxanthine |
| PDE | phosphodiesterase |
| PDEinh | Inactivated variant of phosphodiesterase |
| PPi | pyrophosphate |
Parameter finding
A description of the process to find bond-graph parameters is shown in the folder parameter_finder, which relies on the:
- stoichiometry of system
- kinetic constants for forward/reverse reactions
- If not already, all reactions are made reversible by assigning a small value to the reverse direction.
Here, this solve process is performed in Python.
Original kinetic model
This bond-graph network is largely based on cAMP metabolism of Saucerman et al: Modeling beta-adrenergic control of cardiac myocyte contractility in silico.

