BG_PLC
About this model
This is a bond-graph model of the metabolism of the phospholipase-C beta variant (PLC) and the associated Gq protein in the mammalian cell.
- INPUTS:
- Gq protein (Gq)
- OUTPUTS:
- Change in molar amount of diacylglycerol (DAG), inositol trisphosphate (IP3), calcium (Ca)
- REACTIONS:
- R0: Conversion of DAG into phosphatidylcholine (PC)
- R1: Conversion of IP3 into inositol (Ino)
- R2: PLC complex CaPLC catalysing the splitting of phosphatidylinositol biphosphate (PIP2) into DAG and IP3
- R3: Similar to R2, but with enzyme complex CaGqPLC
- R4: Binding of Ca and PLC
- R5: Binding of Gq and PLC
- R6: Binding of Ca and GqPLC
- R7: Unbinding of Gq from CaGqPLC
- R8: Unbinding of Gq (in G*GDP form) from CaGqPLC
Model status
The current CellML implementation runs in OpenCOR.
Model overview
This model is based on existing kinetic model, where the mathematics are translated into the bond-graph formalism. This describes the model in energetic terms and forces adherence to the laws of thermodynamics.
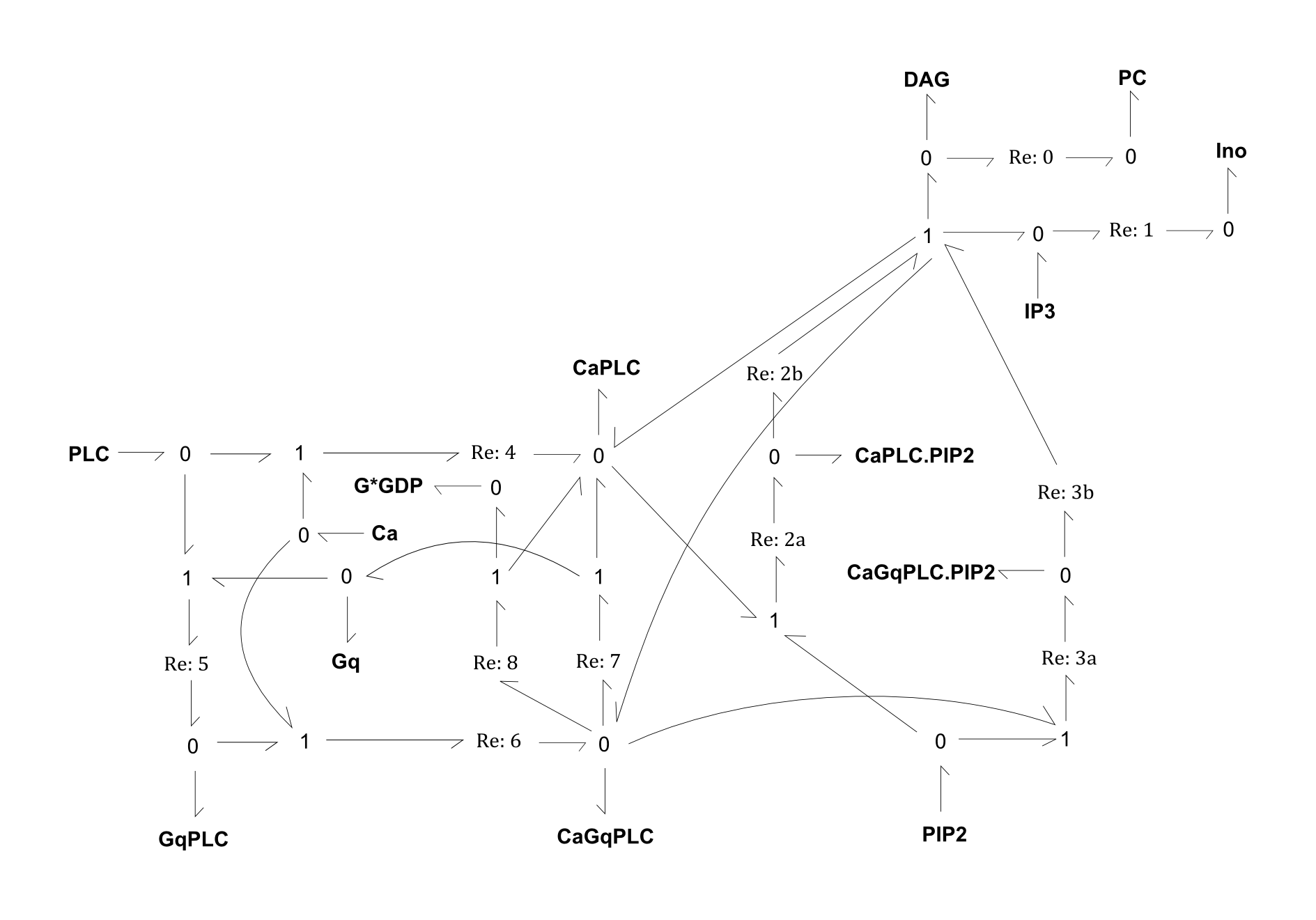
Fig. 1. Bond-graph formulation of the PLC network
For the above bond-graphs, a '0' node refers to a junction where all chemical potentials are the same. A '1' node refers to all fluxes being the same going in and out of the junction.
| Abbreviation | Name |
|---|---|
| PLC | Phospholipase-C (beta variant) |
| Ca | Calcium ion |
| Gq | Gq protein |
| DAG | Diacylglycerol |
| IP3 | Inositol trisphosphate |
| Ino | Inositol |
| PIP2 | Phosphatidylinositol biphosphate |
| PC | Phosphatidylcholine |
Parameter finding
A description of the process to find bond-graph parameters is shown in the folder parameter_finder, which relies on the:
- stoichiometry of system
- kinetic constants for forward/reverse reactions
- If not already, all reactions are made reversible by assigning a small value to the reverse direction.
Here, this solve process is performed in Python.
Original kinetic model
Bhalla and Iyengar: Emergent properties of networks of biological signaling pathways.

