FAIRDO BG example 3.4.cellml
Bond graph example: A simple biochemical reaction
For biochemical systems qij is the molar quantity of a chemical species i in solution in compartment j and uij is its chemical potential. A biochemical reaction in one compartment ('c'), with forward and reverse affinities Af and Ar, is shown below along with its bond graph representation. Note the usual representation for energy storage, but the dissipative reaction term (Rer) now requires explicit dependence on the chemical potential of the reactants and products, rather than depending only on the potential across the dissipation 'resistor'. Therefore, the corresponding Re component must have two ports, in contrast to the standard one-port R component. Flow 1:nodes are therefore needed on either side of the reaction, when multiple species are involved, to provide the two ports.
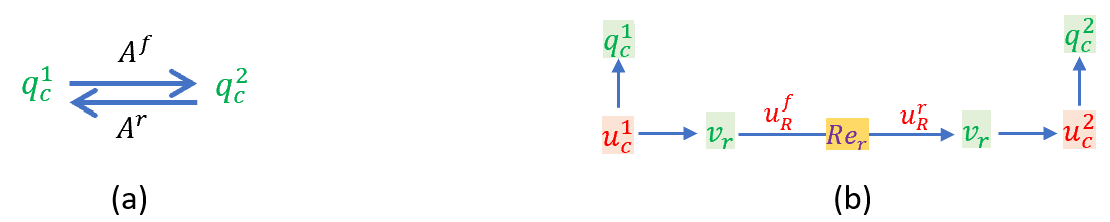
A biochemical reaction where Af and Ar are the forward and reverse affinities (a), and its bond graph representation (b). The 0:nodes represent the chemical potentials ui1 and ui2 associated with the chemical species qi1 and qi2 stored in compartments 1 and 2. The reaction Rer (the dissipative term for a biochemical system) has associated chemical flux vr that depends explicitly on the chemical potentials of the reactant (q1c) and product (q2c) since in this simple case ufR = Af = u1c and urR = Ar = u2c.
The Views Available menu to the right provides various options to explore this model here in the Physiome Model Repository. Of particular interest is the Launch with OpenCOR menu item, which will load the simulation experiment shown below directly into the OpenCOR desktop application.
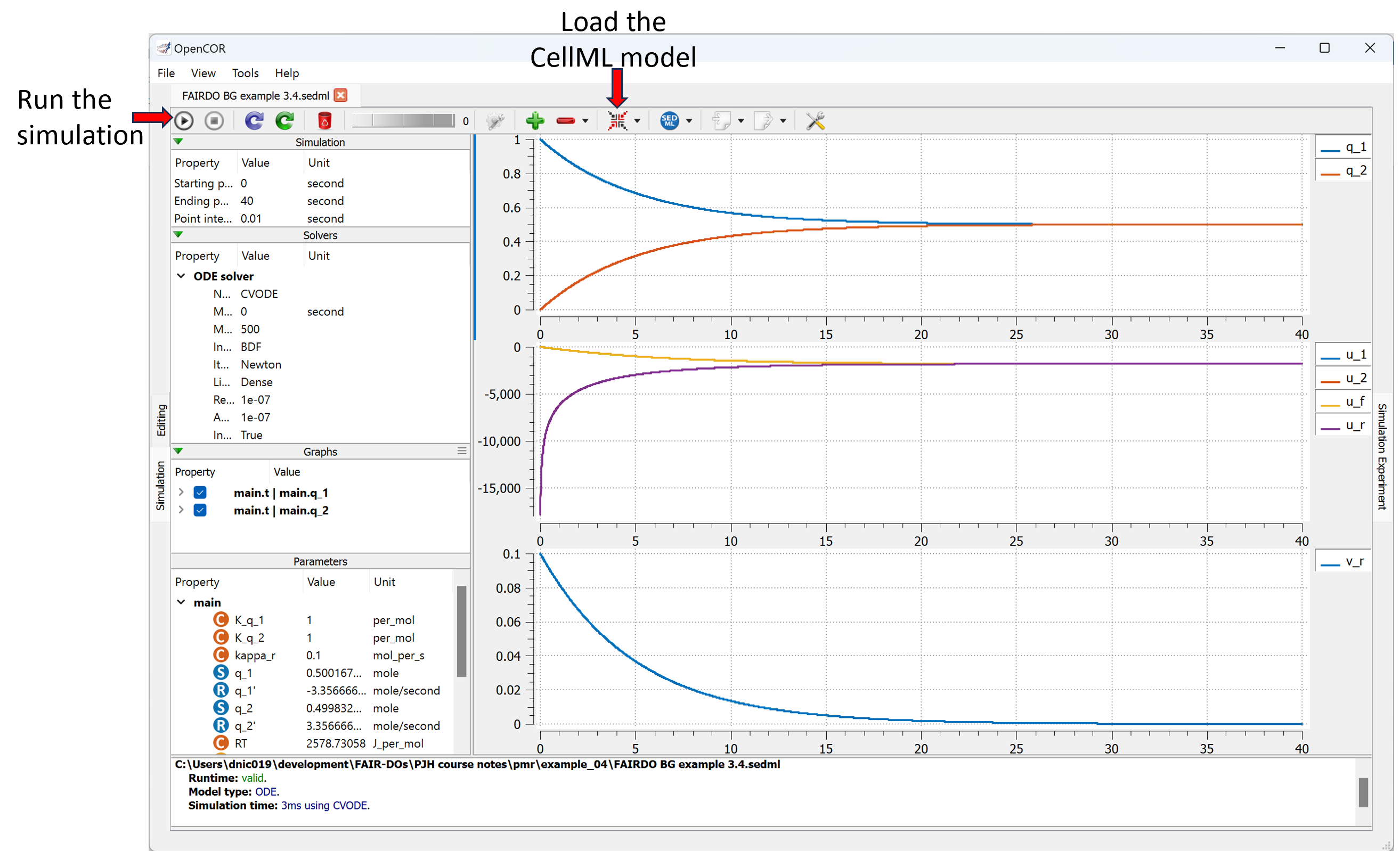
Showing the result of launching the simulation experiment from this exposure in OpenCOR and executing the simulation.

