Location: Dynamics of amino acid metabolism of primary human liver cells in 3D bioreactors @ fbf661b41bf1 / guthke_2006.html
- Author:
- Hanne Nielsen <hnie010@aucklanduni.ac.nz>
- Date:
- 2011-08-23 12:23:52+12:00
- Desc:
- Added HTML file
- Permanent Source URI:
- https://models.cellml.org/w/hnielsen/guthke_2006/rawfile/fbf661b41bf1598575a6b29d3cffad0261edf30d/guthke_2006.html
Model Status
This CellML model runs in OpenCell and COR but does not exactly reproduce published results
Model Structure
The kinetics of 18 amino acids, ammonia (NH3) and urea (UREA) in 18 liver cell bioreactor runs were analyzed and simulated by a two-compartment model consisting of a system of 42 differential equations. The model parameters, most of them representing enzymatic activities, were identified and their values discussed with respect to the different liver cell bioreactor performance levels. The nitrogen balance based model was used as a tool to quantify the variability of runs and to describe different kinetic patterns of the amino acid metabolism, in particular with respect to glutamate (GLU) and aspartate (ASP).
The original paper reference is cited below:
'Dynamics of amino acid metabolism of primary human liver cells in 3D bioreactors.', Guthke R, Zeilinger K, Sickinger S, Schmidt-Heck W, Buentemeyer H, Iding K, Lehmann J, Pfaff M, Pless G, Gerlach JC., 2006 Bioprocess Biosyst Eng, 28(5), 331-40. PubMed ID: 16550345
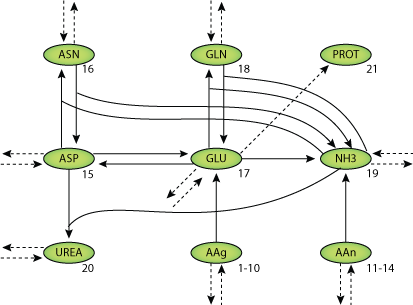
|
| Structure of the model Equations. |

