FCU_adenylylCyclase
About this model
This is a Functional Cell Unit for adenylyl cyclase metabolism in the cardiac cell.
This is a composite module, composed of several individual modules, and merged together in a modular fashion.
- INPUTS:
- Ligands for beta-1 adrenergic and muscarinic receptors.
- OUTPUTS:
- Change in molar amount of cyclic AMP.
Model status
The current CellML implementation runs in OpenCOR.
Model overview
All components are presented in bond-graph form. Components are made by converting an existing kinetic model and translating into bond-graph form.
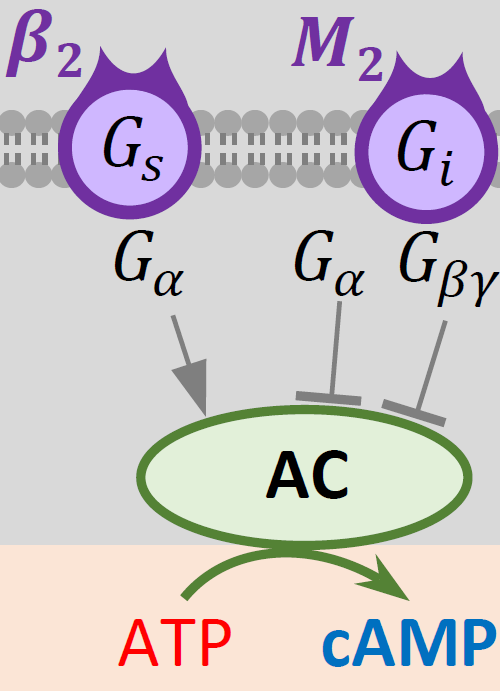
Components of the model.
A description of the process to find bond-graph parameter is shown in the folder parameter_finder, which relies on the:
- stoichiometry of system
- kinetic constants for forward/reverse reactions
- If not already, all reactions are made reversible by assigning a small value to the reverse direction.
Here, this solve process is performed in Python.
Modular description
Components
CellML divides the mathematical model into distinct components, which are able to be re-used in other composite models. These CellML components are:
- beta-1 adrenergic receptor and Gs protein (GPCR-β 1AR)
- muscarinic receptor and Gi Protein (GPCR-M2) (M2)
- cyclic AMP (cAMP)
| Source | Components |
|---|---|
| Saucerman et al. | GPCR-β 1AR, cAMP |
| Iancu et al. | GPCR-M2 |
Each of these blocks is itself a CellML model, complete with bond-graph parameters appropriate for the isolated system.

