SinglePASMC
About this model
| Original publication: | |
|---|---|
| Gosak, Marko, et al (2014): "The influence of gap junction network complexity on pulmonary artery smooth muscle reactivity in normoxic and chronically hypoxic conditions." Experimental physiology 99.1 (2014): 272-285. | |
| DOI: | 10.1113/expphysiol.2013.074971 |
Model status
The current CellML implementation runs in OpenCOR. The results have been validated against the data extracted from the figures in the published Gosak, Marko, et al (2014). We provide the settings used for the figure reproduction with the simulation results shown under Experiments. The model structure can be found in the documentation of Components. The curation process has been summarized in the Model history and Known issues.
Model overview
This workspace holds a CellML encoding of the Gosak, Marko, et al (2014) model. The Gosak, Marko, et al (2014) model describes major elements involved in transplasmalemmal ion exchange and intracellular Ca2+ handling, while the formulism was adapted from the Parthimos et al.(1999) and Koenigsberger et al. (2004).
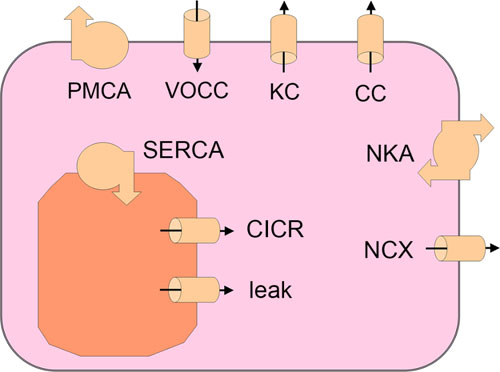
A diagrammatic representation of the Gosak, Marko, et al (2014) model.
Modular description
Components
CellML divides the mathematical model into distinct components, which are able to be re-used. The main CellML components are:
- The major elements involved in transplasmalemmal ion exchange, including:
Ca2 + concentration in the cytosol (c): Cai
Ca2 + concentration in sarcoplasmic reticulum (s): Casr
The dynamics of the cell membrane potential (v): Vm
Each of these blocks is itself a CellML model, which enables us to reuse the various components in future studies and models.
Experiments
Following best practices, this model separates the mathematics from the parameterisation of the model. The mathematical model is imported into a specific parameterised instance in order to perform numerical simulations. The default parameters are defined in Para The parameterisation would include defining the stimulus protocol to be applied.
This workspace encodes single-cell response to KCl stimulation and corresponding simulation results.
Simulation settings
Simulation settings are encoded in SED-ML documents for experiment execution. It is common that we may need to vary experimental settings to obtain data under various conditions. Hence, the full experimental settings are encoded in the simulation scripts. The Python scripts to run simulation and reproduce the figures in the original paper are included under the Simulation/src folder. The runSim.ps1 is used to run the simulation in PowerShell.
Model history
There is no publicly available code for this model.
Known issues
- There are some parameters and initial values need to be confirmed:
- The simulation result is not exactly align with the original data, which could be caused by the parameter settings in Table 1.

