Reed, Nijhout, Sparks, Ulrich, 2004
Model Status
This model runs in COR and OpenCell and the units are consistent throughout. The model recreates the published results (Figures 4A and 4B), and is able to simulate other figures if the Metin and 5mTHF values are varied.
Model Structure
ABSTRACT: Building on the work of Martinov et al. (2000), a mathematical model is developed for the methionine cycle. A large amount of information is available about the enzymes that catalyse individual reaction steps in the cycle, from methionine to S-adenosylmethionine to S-adenosylhomocysteine to homocysteine, and the removal of mass from the cycle by the conversion of homocysteine to cystathionine. Nevertheless, the behavior of the cycle is very complicated since many substrates alter the activities of the enzymes in the reactions that produce them, and some can also alter the activities of other enzymes in the cycle. The model consists of four differential equations, based on known reaction kinetics, that can be solved to give the time course of the concentrations of the four main substrates in the cycle under various circumstances. We show that the behavior of the model in response to genetic abnormalities and dietary deficiencies is similar to the changes seen in a wide variety of experimental studies. We conduct computational "experiments" that give understanding of the regulatory behavior of the methionine cycle under normal conditions and the behavior in the presence of genetic variation and dietary deficiencies.
The original paper reference is cited below:
A mathematical model of the methionine cycle, Michael C. Reed, H. Frederik Nijhout, Rachel Sparks, and Cornelia M. Ulrich, 2004, Journal of Theoretical Biology, 226, 33-43. PubMed ID: 14637052
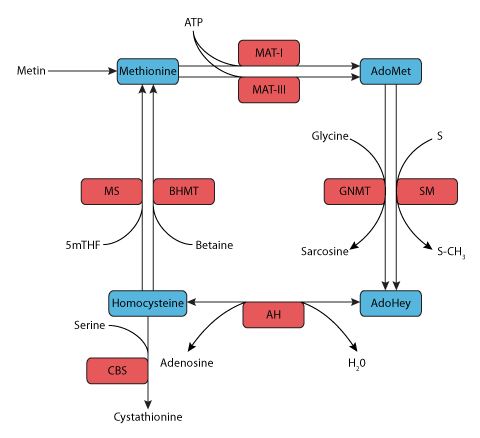 |
| Components of the methionine metabolism modelled by Reed et al.. The main metabolites are shown in blue boxes and the enzymes in red boxes. |

